Press release -
Indicator molecule determined for mitochondrial toxicity arising from side effects of medicine -- High hopes of application to a new method of toxicity assessment in drug development -- Kindai University
A research group consisting of Professor Kei Zaitsu (Kindai University Faculty of Biology-Oriented Science and Technology, Kinokawa-shi, Wakayama prefecture), Akira Iguchi (chief scientist at National Institute of Advanced Industrial Science and Technology, Tsukuba-shi, Ibaraki prefecture), and Yui Hibino (chief scientist at Mitsubishi Tanabe Pharma Corporation, Fujisawa-shi, Kanagawa prefecture) has, through metabolomic analysis and bioinformatics, revealed an indicator molecule which can be used to determine the mechanism of mitochondrial toxicity, a cause of drug-induced liver disease (a side effect of medicine). These results could be applied to the formation of a new toxicity assessment method in the drug development field.
These results were published in the international journal of toxicology, “Toxicology and Applied Pharmacology” at 8:45AM on Thursday, December 1st, 2022 (Japan time).

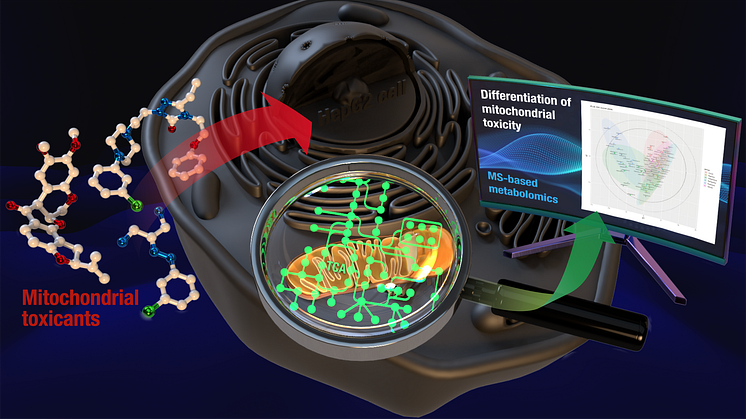
An image of mitochondrial toxicity mechanism determination through metabolomic analysis and bioinformatics
1. Key Points
- Conducted metabolomic analysis on human hepatoma cell lines which were exposed to mitochondrial toxic compounds
- Utilized bioinformatics to identify the indicator molecule which determines the mechanism of mitochondrial toxicity
- These results could be applied to a new toxicity assessment method in the drug development field
2. Research Background
Drug-induced liver disease is a liver inflammation which occurs as a side effect of medicine. When found in patients taking a certain medicine, it can lead to discontinuation of sales or development. Therefore, it is important to accurately predict the risk of drug-induced liver disease.
One major cause of drug-induced liver disease is mitochondrial toxicity. The mitochondrion is a subcellular organelle and is often referred to as the “energy factory” of cells. Mitochondrial toxicity is a characteristic that impairs the function of the mitochondria.
Mitochondrial toxicity in medicine has been assessed by exposing the medicine to cultured cells, and then observing the configuration of the mitochondria or monitoring the cell’s oxygen consumption. However, it is essential to determine the mechanism of mitochondrial toxicity in order to predict the risk of drug-induced liver disease, and therefore existing assessment methods were insufficient. A method which utilizes molecular profiling must be established in order to accurately determine the mechanism of mitochondrial toxicity. Said research group has been developing a new technique combining metabolomic analysis and bioinformatics.
3. Content
Aiming to identify the guiding molecules for determining the mechanism of mitochondrial toxicity, the research group exposed 4 chemical compounds (rotenone, CCCP, nefazodone, perhexiline) with different toxicity mechanisms to human hepatoma cell lines (HepG2 cells). They then performed metabolomic analysis and identified 125 metabolites. After analysis with bioinformatics, they revealed that the molecular profile of these 125 components differ greatly when the toxicity mechanism is different.
They also utilized bioinformatics to focus on 50 components and performed machine learning tests to find that these 50 components could act as guiding molecules for mitochondrial toxicity mechanisms. This signifies the possibility of using the guiding molecules to identify the mechanism of mitochondrial toxicity.
These research results are a breakthrough in mitochondrial toxicity mechanism identification. There are high hopes of applying the guiding molecules found in this research to establish a new toxicity assessment method in the field of drug development.
4. About Publication
Journal name:
Toxicology and Applied Pharmacology (Impact factor: 4.46@2021)
Research paper title:
Preliminary study to classify mechanisms of mitochondrial toxicity by in vitro metabolomics and bioinformatics
Authors:
Yui Hibino1, Akira Iguchi2, Kei Zaitsu3*
*Corresponding author
Affiliation:
1. Mitsubishi Tanabe Pharma Corporation
2. National Institute of Advanced Industrial Science and Technology
3. Kindai University Faculty of Biology-Oriented Science and Technology

